Abstract
- The Illumina 5-base solution provides a streamlined, end-to-end NGS library prep and analysis workflow for simultaneous methylation profiling and high-accuracy genetic variant calling.
- An Illumina-engineered enzyme is used to convert methylated cytosines (5mC) to thymine while preserving unmethylated cytosines, which maximizes alignment and enables a 5-base readout of A, G, C, T, and 5mC from each sequencing read.
- The workflow consists of less than one day of library preparation followed by standard Illumina sequencing and ultra-fast DRAGEN dual-omic secondary analysis in less than one hour and is compatible with targeted enrichment.
- Early testing demonstrates that the Illumina 5-base solution offers methylation profiling with accuracy on par with on-market methylation technologies and simultaneous high-accuracy genetic variant calling.
Introduction
DNA molecules contain multiple layers of information, including genetic sequences and epigenetic modifications, making them inherently multiomic. In addition to the four canonical nucleotides, there is a fifth base: methylated cytosine (Figure 1). The methylated cytosines are epigenetic tags that act as dynamic molecular signals to modify gene expression in response to environmental, developmental, and cellular factors. Much like genetic variants provide insight into health states, aberrant methylation signatures can serve as indicators of disease. Layering genetic and epigenetic information together can enhance resolution and sensitivity relative to single-omic studies alone, revealing new biological insights.
.png)
Figure 1: DNA molecules consist of 5 bases: A, C, G, T, and methylated C
Commercially available methylation profiling methods such as bisulfite or EM-seq convert unmethylated cytosines to thymine, removing the majority of cytosines from the library and resulting in a low-complexity genome that is challenging to sequence and align. Methylated cytosines are then inferred from the remaining unconverted cytosines. While these technologies have been fundamental tools in shaping our current understanding of DNA methylation, they lack sufficient accuracy to also enable high-resolution genetic variant calling. We envisioned that a solution offering detection of both variant types from the same workflow would greatly accelerate discoveries.
In that context, we introduce an innovative 5-base solution that enables methylation profiling and genetic variant calling in a fast, simple, and comprehensive workflow relative to alternative approaches (Figure 2).
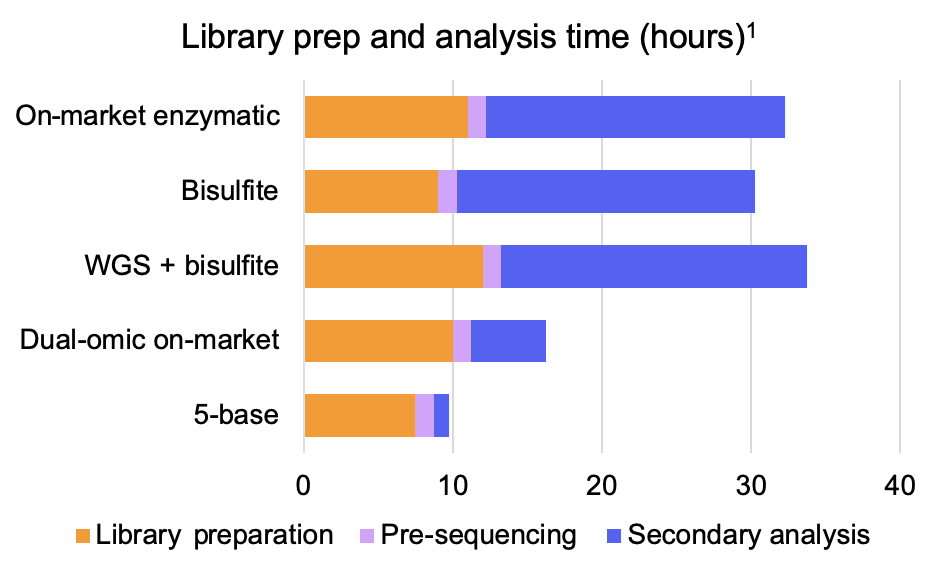
Figure 2: The Illumina 5-base solution offers a shorter turnaround time compared to alternate methylation profiling or dual-omic library prep and analysis options
1. Dual-omic on-market: Füllgrabe et al. 2023; On-market enzymatic: Vaisvila et al. 2021 (library prep) and Bismark; Bisulfite: xGen Methyl-Seq DNA Library Prep Kit (assessed March 2025); WGS: Illumina DNA Prep and DRAGEN pre-sequencing steps include quantification, normalization, and pooling
A novel solution
At the core of the Illumina 5-base solution is a novel, bespoke enzyme that was engineered in-house at Illumina for selective and direct conversion of methylated cytosines to sequencer-ready thymine (Figure 3). Millions of protein variants were screened, over iterative generations, to identify a protein with the highest selectivity for methylated cytosine and efficiency such that the reaction performs optimally in only 30 minutes.
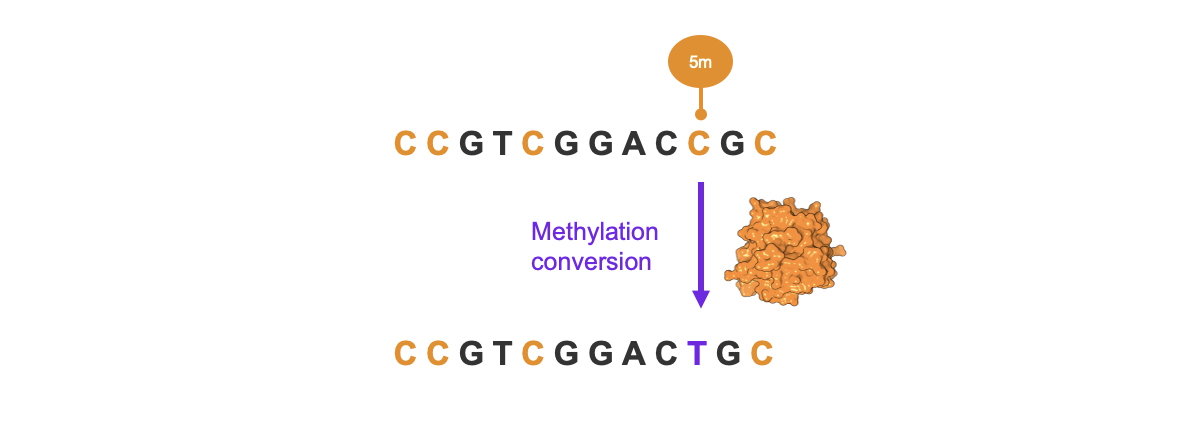
Figure 3: Novel enzyme engineered for highly selective conversion of methylated cytosine (5mC) to thymine
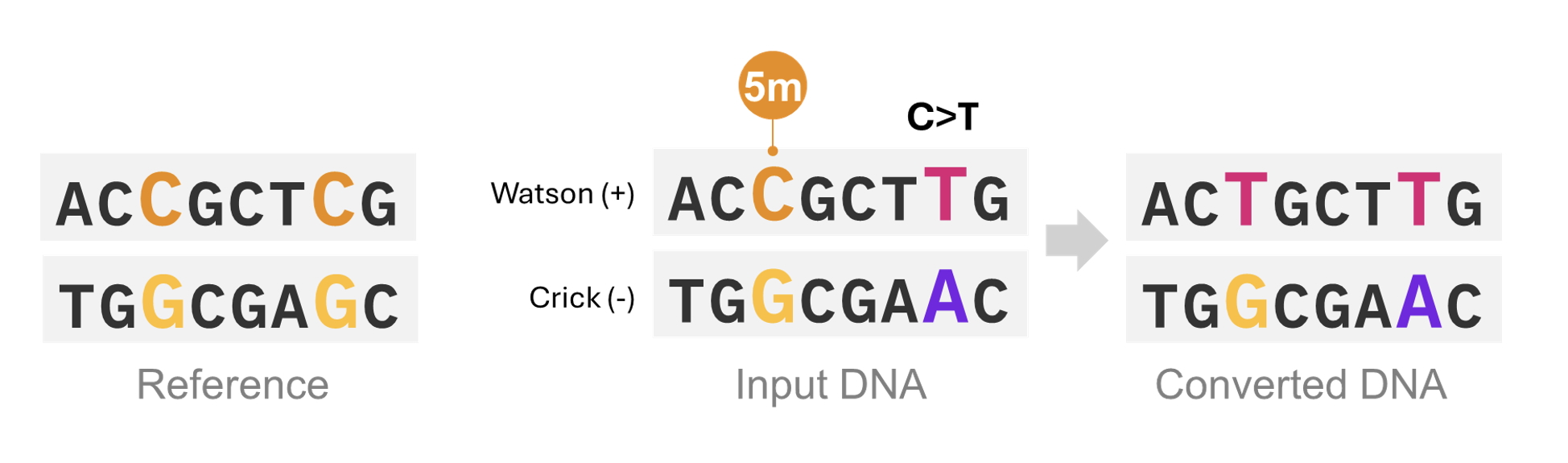
Figure 4: Methylated C versus C>T variants are resolved from DNA duplex
How it works
The methyl cytosine-conversion reaction is a simple, integrated step within a high-efficiency ligation-based library preparation workflow. The workflow begins with extracted, sheared genomic DNA (gDNA) or cell-free DNA (cfDNA), which then undergoes end-repair, A-tailing, and adapter ligation. Next, the DNA is denatured and incubated with the 5-methyl cytosine conversion enzyme for direct conversion of methylated cytosines in a single reaction in less than an hour. After conversion, samples are amplified by PCR and bead purified for sequencing on standard lllumina sequencing platforms. The library preparation is highly streamlined and can be completed in less than eight hours. The conversion chemistry is also compatible with targeted capture enrichment for high-depth applications, including UMI-based error correction.
Once sequencing is complete, the alignment, methylation annotation, and variant calling can be performed with a single push-button analysis pipeline in as little as an hour. The results can be further processed with interpretation software, such as Illumina Connected Multiomics and Emedgene.
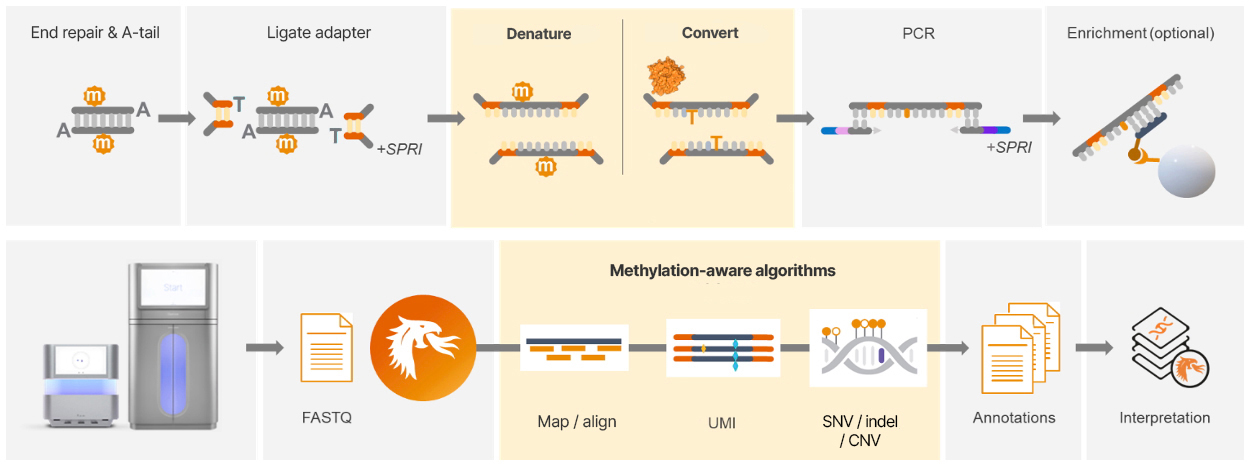
Figure 5: The 5-base workflow
High-accuracy methylation profiling with less sequencing
In benchmarking studies, the 5-base solution provides methylation profiling with comparable accuracy to gold standard methylation conversion methods. The chemistry achieves high conversion rates of methylated cytosines and very low off-target conversion of unmethylated cytosines based on evaluation with methylated and unmethylated pUC and lambda control genomes (Figure 6A). Methylation levels measured with the 5-base solution across CpG islands in human genomes are highly consistent with values obtained from EM-seq conversion. The high mappability of 5-base libraries from converting only the methylated cytosines results in more CpG sites covered per sequencing depth (Figure 6C), which translates to a higher discovery power with the 5-base solution.
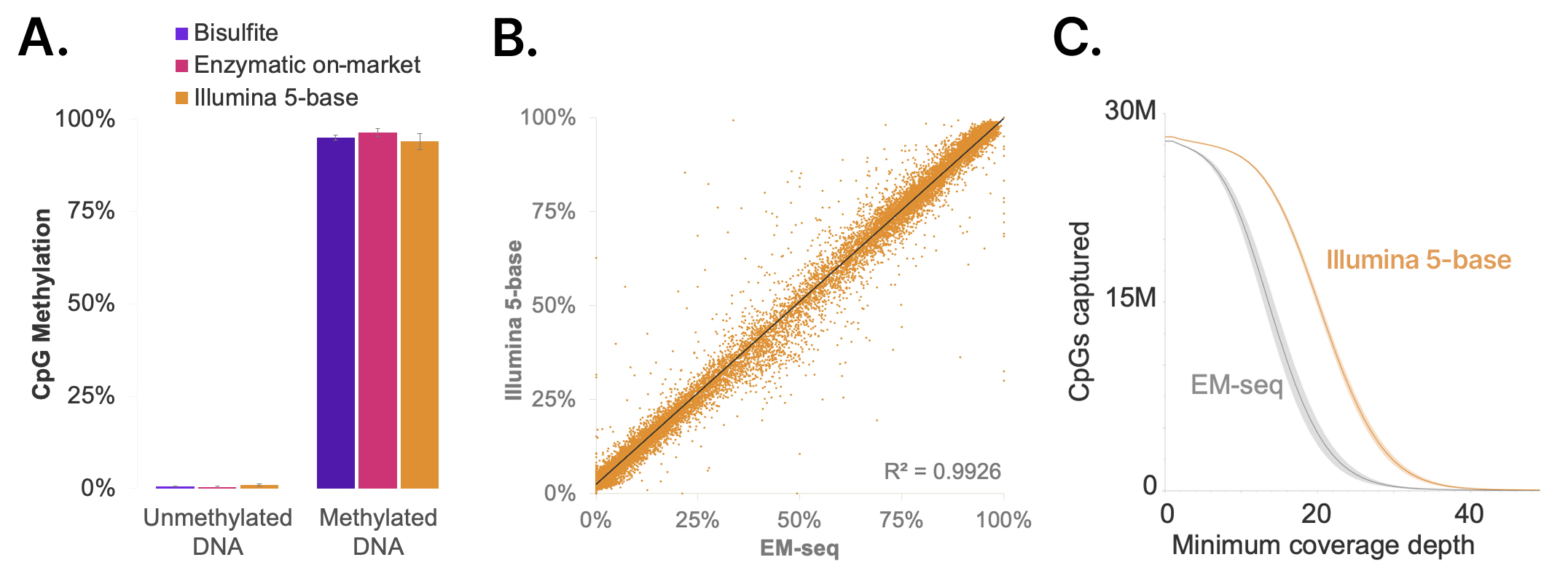
Figure 6: High-accuracy methylation detection with fewer sequencing reads compared to on-market gold standard methyl-seq chemistries
(A) The Illumina 5-base solution accurately and specifically converts methylated control DNA (unmethylated lambda: 0.5%, 0.4%, 0.9%; methylated pUC19: 95.0%, 96.4%, 94.7% for Bisulfite, EM-seq v1, and 5-base conversion methods performed with Illumina ligation-based library prep) (B) Illumina 5-base is highly correlated with EM-seq when comparing average methylation in CpG islands of NA12878 reference DNA (C) Number of CpGs captured from 500 million sequenced read pairs with Illumina 5-base versus EM-seq v1 conversion. 27 million CpG sites have 10× coverage with 5-base compared to 22 million CpGs with EM-seq.
Simultaneous detection of genetic variants with high sensitivity
Methylation chemistries that convert unmethylated cytosine to thymine pose challenges for genetic variant detection and typically require that standard sequencing libraries be processed separately if genetic variant information is desired. The Illumina 5-base solution eliminates the need for two separate library preparations by enabling high-resolution variant detection at all four base positions (A, T, G, and C) in addition to methylated cytosine detection within the same sequencing read. When analyzed with DRAGEN applications available in Illumina BaseSpace Sequencing Hub or Illumina Connected Analytics, the 5-base solution delivers high-accuracy small variant calling—for both germline and somatic variants (Figures 7A, 7B). Additionally, the technology can be combined with targeted capture enrichment for detection of low-frequency variants from blood, fresh-frozen tissue, or cfDNA (Figure 7B).
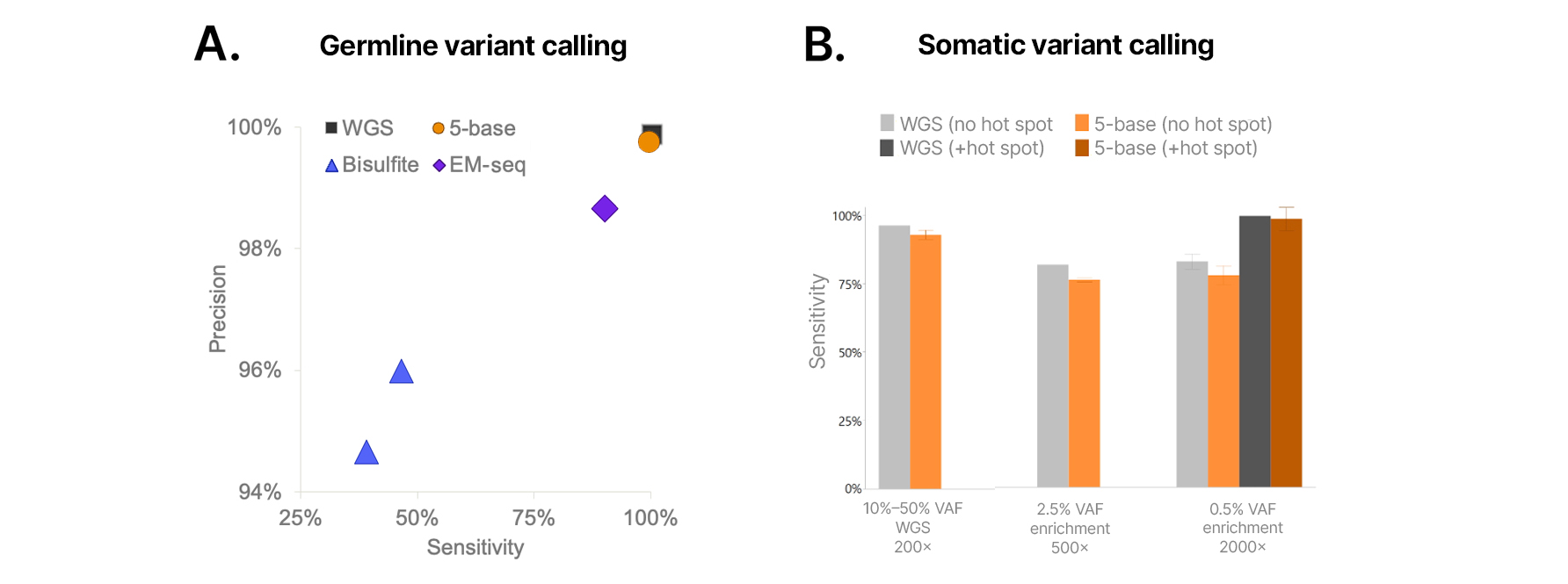
Figure 7: DNA variant calling performance is comparable to standard WGS
(A) Germline SNV calling accuracy of the Illumina 5-base solution for NA12878 (EM-seq and Bisulfite processed by Bis-SNP, 5-base and WGS by DRAGEN) is comparable with WGS. (B) Low variant allele frequency (VAF) SNV detection with enrichment compared to standard WGS with enrichment and UMI collapsing
One assay with dual-omic insights empowers discovery
The Illumina 5-base solution is a novel approach for harnessing genetic and epigenetic insights from a single workflow that we anticipate will enhance discovery power across broad research applications, including cancer and rare disease. The simultaneous reporting of genome and methylome status on a single read can be leveraged to link pathogenic mutations to germline allele-specific methylation (Figure 8A). We also demonstrate that the 5-base resolves the case by dual-omic interrogation of genome and methylation signatures obtained from a single sample preparation (Figure 8B). At Illumina, we are continuing to explore how combining dual-omic insights will drive new discoveries across a wide range of exciting applications. We anticipate that the 5-base solution will be available commercially in early 2026.
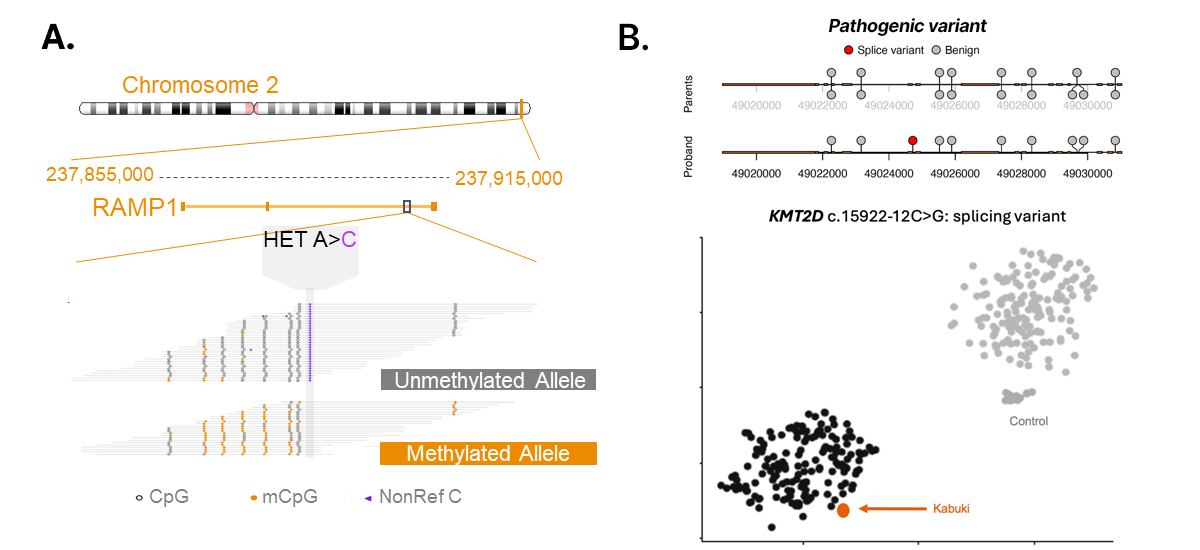
Figure 8: Multiomic insights enhance resolution and empower discovery
(A) Simultaneous read-level reporting of genome and methylome captures germline allele-specific methylation on one allele. A missense mutation and absence of methylation is observed on the other allele, signatures that cannot be resolved by standard WGS alone. (B) Processing of a DNA sample from a rare disease research participant with 5-base technology. Small variant detection through Illumina DRAGEN germline analysis identified a KMT2D mutation. Methylation data was processed through an epigenetic disease classifier algorithm developed by EpiSign. Taken together, the mutation and epigenetic signature classify the disease as Kabuki syndrome.