Rare Disease Whole-Genome Sequencing
The most comprehensive test for rare disease1–8
Whole-genome sequencing offers the highest likelihood of finding a diagnosis for rare genetic disease10
Benefits of WGS for Rare Disease
Whole-genome sequencing (WGS) for rare disease offers three key advantages over other genetic testing methods:
- Potential for greater diagnostic yield10
- Improved operational efficiency
- Decreased cost3,6
Especially important for rare disease cases, whole-genome sequencing is the most comprehensive test for detecting multiple variant types in a single assay.1–8 In a large, randomized controlled trial, the median time to diagnosis in neonatal intensive and pediatric intensive care patients was 13 days with WGS, compared to 107 days with standard testing.9
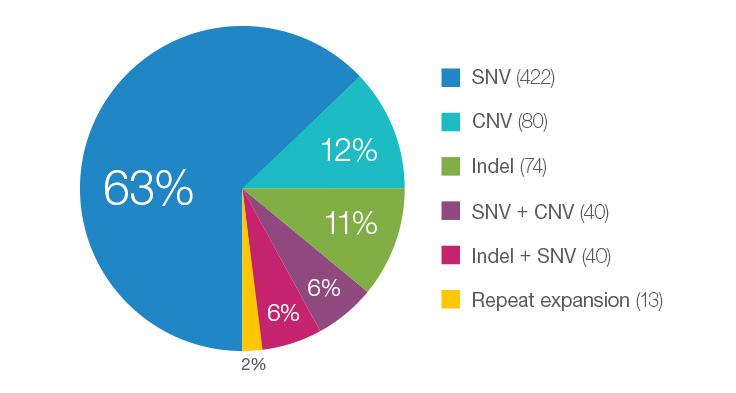
Many Different Variant Types are Pathogenic for Rare Disease Cases
WGS testing performed in the Illumina Clinical Services Laboratory represents individuals enrolled in disease-specific clinical trials or as part of philanthropic efforts. As such, the percentage represented here may not be typical of that seen in a standard laboratory. This data is based on 669 total cases.
WGS Can End Rare Disease Diagnostic Odysseys
Whole-genome sequencing for rare disease has the power to help doctors diagnose genetic diseases quickly, helping families avoid long diagnostic odysseys. Of all genomic testing methods, WGS offers the highest likelihood of finding a diagnosis.10 It provides the highest coverage of the human genome, not only in regions not covered by other methods, but even within regions targeted by other methods.11,12 This increased coverage at first-line usage has been shown to reduce the need for unnecessary iterative tests and reduce the length of stay in the NICU.13,14
WGS can also impact patient care. A change in management has been reported in 49–75% of pediatric outpatients who received a diagnosis by WGS.15,16
Expanding Access to WGS for Rare Diseases
By publishing best practices, the Medical Genome Initiative aims to expand access to high-quality WGS for genetic disease diagnosis.
Read ArticleWGS Identifies Causes of Rare Disease

The Key To Implementing Genomes: Collaboration
Learn how the University of Utah and ARUP Laboratories have collaborated to deliver answers faster with genome sequencing.
Watch Video
Taking a Genome-First Approach
The team at HudsonAlpha have been innovators in integrating genome sequencing into medicine. Hear why genomes were central to their vision.
Watch VideoAmerican College of Medical Genetics and Genomics (ACMG) Highlights
Learn about evidence-based guidelines for exome and genome sequencing for pediatric patients with congenital anomalies or intellectual disability/developmental delay.
View FlyerGenome-Wide Sequencing as a Diagnostic Test
Learn about recent society statements on genome-wide sequencing.
View TableHear from Experts on Rare Disease WGS

Standardization of Clinical WGS for Rare Disease
Dr. Christian Marshall of The Hospital for Sick Children explains how laboratory and clinical best practices can enable whole-genome sequencing for genetic disease diagnosis.
WGS and the End of the Diagnostic Odyssey
Dr. Vandana Shashi of Duke University and Kimberly LeBlanc of the Undiagnosed Diseases Network discuss how WGS can short-circuit the diagnostic odyssey for patients with rare disease.

Rapid Whole-Genome Sequencing of Critically Ill Children
Dr. Shimul Chowdhury explains how rapid WGS can help pinpoint the causes of rare disease in children.
Featured Rare Disease WGS Publications
100,000 Genomes Pilot on Rare Disease Diagnosis
Pilot study of WGS in a national health care system showed an increase in diagnostic yield across a range of rare diseases.
Effect of WGS on Clinical Management of Infants with Suspected Genetic Disease
Access to WGS doubled the proportion of patients with a precision diagnosis and a change of clinical management.
Integration of WGS into a Healthcare Setting
High diagnostic rates across multiple clinical entities in 3219 rare disease patients analyzed by WGS.

Performance, Utility, and Implementation of WGS in a Clinical Setting
This collection of publications show WGS is the most comprehensive test for detecting multiple variant types in a single assay and that it can provide a genetic diagnosis faster than current genetic testing approaches in acutely ill infants and undiagnosed pediatric outpatients.
View Summary BookletReferences
- Lionel AC, Costain G, Monfared N, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med. 2017; Aug 3. doi: 10.1038/gim.2017.119.
- Sanghvi RV, Buhay CJ, Powell BC, et al. Characterizing reduced coverage regions through comparison of exome and genome sequencing data across 10 centers. Genet Med. 2018;20(8):855-866. doi:10.1038/gim.2017.192
- Dolzhenko E, van Vugt JJFA, Shaw RJ, et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017;27(11):1895-1903. doi:10.1101/gr.225672.117
- Gross A, Ajay SS, Rajan V, et al. Copy number variants in clinical WGS: deployment and interpretation for rare and undiagnosed disease. Genetic Med. 2019;21(5):1121-1130.
- Alfares A, Aloraini T, Subaie LA, et al. Whole-genome sequencing offers additional but limited clinical utility compared with reanalysis of whole-exome sequencing. Genet Med. 2018;20(11):1328-1333. doi:10.1038/gim.2018.41
- Lindstrand A, Eisfeldt J, Pettersson M, et al. From cytogenetics to cytogenomics: whole genomes sequencing as a first-line test comprehensively captures the diverse spectrum of disease-causing genetic variation underlying intellectual disability. Genome Med. 2019;11(1):68.
- Chen, X, Sanchis-Juan, A., French, CE et al. Spinal muscular atrophy diagnosis and carrier screening from genome sequencing data. Genet Med. 2020; 22:945–953. https://doi.org/10.1038/s41436-020-0754-0
- Chen X, Schulz-Trieglaff O, Shaw R, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32(8):1220–1222. http://doi.org/10.1093/bioinformatics/btv710.
- Petrikin JE, Cakici JA, Clark MM, et al. The NSIGHT1-randomized controlled trial: rapid whole-genome sequencing for accelerated etiologic diagnosis in critically ill infants. NPJ Genom Med. 2018;3:6. Published 2018 Feb 9. doi:10.1038/s41525-018-0045-8
- Clark, M.M., Stark, Z., Farnaes, L. et al. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. npj Genomic Med. 2018;3:16. https://doi.org/10.1038/s41525-018-0053-8
- Meienberg J, Bruggmann R, Oexle K, Matyas G. Clinical sequencing: is WGS the better WES? Hum Genet. 2016;135(3):359-362. doi:10.1007/s00439-015-1631-9
- Belkadi A, Bolze A, Itan Y, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci U S A. 2015 Apr 28;112(17):5473-8. doi: 10.1073/pnas.1418631112
- Farnaes L, Hildreth A, Sweeney NM, et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom Med. 2018;3:10. Published 2018 Apr 4. doi:10.1038/s41525-018-0049-4
- Stark Z, Lunke S, Brett GR, et al. Meeting the challenges of implementing rapid genomic testing in acute pediatric care. Genet Med. 2018;20(12):1554-1563. doi:10.1038/gim.2018.37
- Scocchia A, Wigby KM, Masser-Frye D, et al. Clinical whole genome sequencing as a first-tier test at a resource-limited dysmorphology clinic in Mexico. NPJ Genom Med. 2019;4:5. Published 2019 Feb 14. doi:10.1038/s41525-018-0076-1
- Bick D, Fraser PC, Gutzeit MF, et al. Successful application of whole genome sequencing in a medical genetics clinic. J Pediatr Genet. 2016;6(2):61-76.