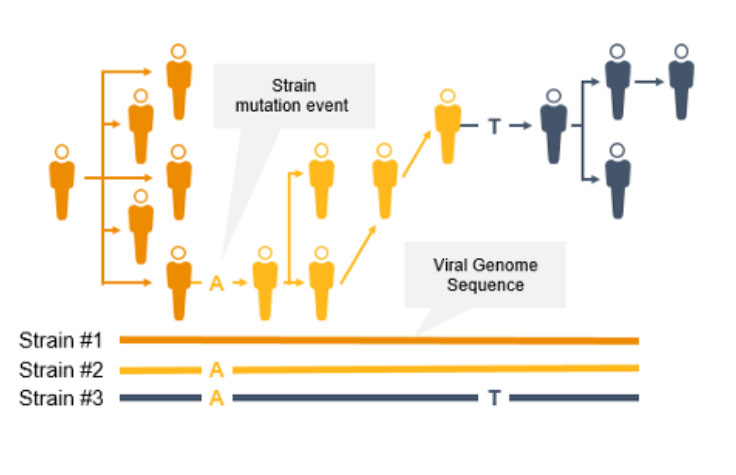
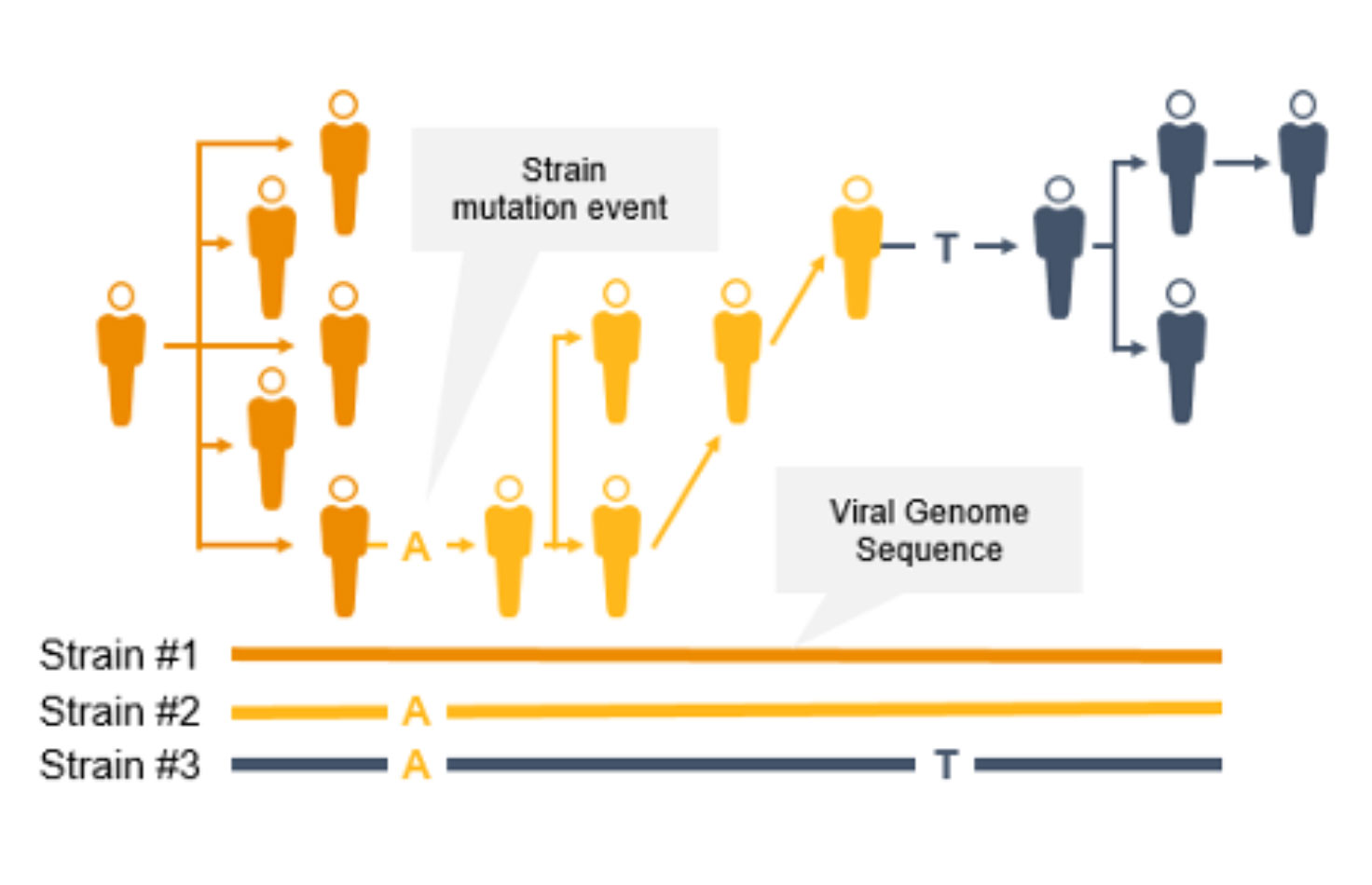
Genomic Surveillance of Infectious Diseases

Identify & Track Infectious Disease Threats
Genomic surveillance with next-generation sequencing (NGS) can track infectious disease transmission and identify novel strains of coronavirus and other emerging pathogens. With near complete sequence data of pathogen genomes, we can implement effective infectious disease surveillance strategies to help prevent further transmission and infection.
Infectious disease surveillance with NGS can:
- Track the transmission routes of infectious diseases globally
- Determine how quickly pathogens such as coronaviruses are mutating as they spread
- Identify novel and known mutations in pathogens like coronavirus, influenza A & B and more
- Study infectious disease therapy resistance
- Investigate vaccine evasion mechanisms
Genomic surveillance helps public health officials track the path of an epidemic, perform contact tracing, determine the rate of pathogen evolution, and understand if a pathogen is changing in ways that could impact diagnostic or therapeutic effectiveness.
Detect & Characterize Coronavirus Mutations
As the coronavirus mutates, new variants evolve such as the Alpha (B.1.1.7, UK), Beta (B.1.351, S. Africa), Gamma (P1, Brazil), Delta (B.1.617.2, India) and Omicron (B.1.1.529, S. Africa) variants. Surveillance and monitoring are critical because mutations can result in greater transmissibility or infectiousness. These coronavirus mutations can potentially make vaccines less effective/protective or even evade test diagnosis.
NGS is a valuable technology for genomic surveillance of infectious diseases such as COVID-19. Not only can it track the prevalence of coronavirus mutant strains, NGS can also identify novel coronavirus mutations, unlike PCR technology. Using NGS, scientists can detect low-frequency minority variants and multiple polymorphisms as well as novel variants.
Genome-wide strain typing with NGS can help epidemiologists identify and characterize mutations quickly to prevent further spread. Strain-level tracing can support identification of outbreak clusters and transmission routes.
In contrast, PCR is designed to detect specific regions of the pathogen genome; it will not identify new mutations across rapidly evolving pathogen genomes. Furthermore, PCR performance can suffer if mutations occur in the primer or probe binding regions.
An Outbreak Anywhere Can Become an Outbreak Everywhere
Monkeypox virus (MPXV) is a large, double-stranded DNA virus first identified in 1958. Historically, the monkeypox virus has caused sporadic human outbreaks in central and west Africa, with a mortality rate between 1% and 10%[1]. In the spring of 2022, outbreaks were observed in non-endemic countries of Europe, North America, and Southeast Asia. Public health surveillance efforts are underway in these countries to prevent the further spread of the monkeypox virus.
NGS is a valuable technology for genomic surveillance of infectious diseases such as monkeypox. Not only can NGS track the prevalence of mutant strains, but it can also identify novel mutations. As a result, scientists can now employ NGS-based technologies to detect low-frequency minority variants, multiple polymorphisms, and novel variants with speed and accuracy in a high-throughput format. These factors are vital in helping epidemiologists identify and characterize outbreak clusters, transmission routes, and mutations to prevent further spread.
Illumina is proud to have supported the global expansion of NGS capacity and capabilities for public health during the COVID pandemic. These resources can now be used for the surveillance of pathogens such as monkeypox virus, ebola virus, mycobacterium tuberculosis, and many others.
Compare NGS Pathogen Surveillance Methods
| Testing Needs | Amplicon | Target Enrichment | Shotgun Metagenomics |
|---|---|---|---|
| Speed & Turnaround Time | |||
| Scalable & Cost-Effective | |||
| Identify Novel Pathogens | |||
| Track Transmission | |||
| Detect Mutations | |||
| Identify Co-Infections & Complex Disease | |||
| Detect Antimicrobial Resistance |
Adequately meets laboratory testing needs
Partially meets laboratory testing needs
More About Infectious Disease Surveillance Sequencing Methods
Amplicon Sequencing
Detect and fully characterize a known virus. Whole genome sequencing via an amplicon approach is ideal for known viruses with small genomes. This method involves analyzing genomic regions of interest with ultra-deep sequencing of PCR amplicons.
Target Enrichment Sequencing
Detect and characterize coronaviruses, flu viruses, and other pathogenic organisms, as well as associated antimicrobial resistance alleles. These insights can help public health officials monitor outbreaks and optimize infection control strategies. This method captures genomic regions of interest via hybridization to target-specific probes.
Shotgun Metagenomics
Comprehensively sequence all organisms in a given sample and identify novel, or emerging pathogens such as mokeypox virus. This NGS method can help accelerate outbreak investigations and support development of new laboratory tests.
Surveillance of Infectious Disease through Wastewater Sequencing
Learn how to use wastewater sequencing to monitor SARS-CoV-2 variants and other respiratory viruses in the community.
Read Application NoteInfectious Disease Surveillance Webinars
NGS for SARS-CoV-2 Detection and Surveillance
NGS has broad applicability for responding to the SARS-CoV-2 pandemic, from initial detection and characterization to monitoring, surveillance, and diagnostic detection.
View WebinarRole of Genomics in Global COVID-19 Surveillance
Join change-makers, thought leaders, and industry shapers for a discussion about the role of genomics in infectious disease surveillance.
View WebinarTargeted Enrichment for Disease and AMR Detection
Earlier and more comprehensive detection of infectious pathogens and antimicrobial resistance (AMR) markers can help optimize patient management and treatment.
View WebinarFeatured Products

COVIDSeq Family
COVIDSeq Assay and COVIDSeq Test are amplicon-based next-generation sequencing (NGS) assays designed to help public health labs identify novel strains of SARS-CoV-2.

iSeq 100
The iSeq 100 system leverages the speed and affordability of complementary metal-oxide-semiconductor (CMOS) technology and the accuracy of sequencing by synthesis (SBS) chemistry.

NextSeq 2000
Groundbreaking benchtop sequencers allow you to explore new science across a variety of current and emerging applications, with higher efficiency and fewer restraints.
Podcast: Genomic Surveillance & Testing for SARS-CoV-2
Christopher Mason from Weill Cornell Medicine describes his multifaceted approach to the urgent need for COVID-19 testing and surveillance. He also discusses his recent Nature paper involving use of omics methods to study COVID-19 host responses and drug interactions.
Preventing the Next Pandemic
Up to 75% of new or emerging infectious diseases are estimated to have zoonotic origins.1,2 We now know that zoonotic reservoirs are significant in the spread of pathogens. With NGS, it is possible to screen reservoir animals such as bats to predict and prevent viral pathogen outbreaks.
Infectious disease surveillance with NGS-based target enrichment or metagenomics can help us understand interspecies transmission, how zoonotic diseases emerge, spread and become resistant to common therapies, and enable us to better contain, treat and prevent disease outbreaks.
Targeted enrichment sequencing can be scaled up to monitor zoonotic pathogens, while genomic surveillance with metagenomics allows for unbiased, culture-free detection and identification of a broad number of pathogens. Metagenomics also helps us understand the relationship between our microbiome and pathogen interactions, which is important when designing measures to control infectious diseases.
Learn more about:
Target EnrichmentMetagenomic Sequencing
More Infectious Disease Surveillance Applications
Coronavirus Surveillance
NGS provides unbiased identification of new coronavirus strains. Illumina offers rapid detection of SARS-CoV-2 coronavirus mutations to meet the demands for efficient sequencing needs.
Foodborne Disease Surveillance
Microbial whole-genome sequencing can access all the genetic variant information of a pathogen as it spreads from food to humans. The more variants we can describe, the higher the probability of finding the sources of infections and routes of transmission.
Viral Surveillance
Target enrichment uses hybridization to capture genomic regions of interest. This method can provide the high sensitivity needed to detect a virus and provide information about its epidemiology and evolution.
Respiratory Infection Surveillance
These panels offer targeted enrichment sequencing solutions for respiratory pathogen and respiratory virus detection and characterization, including COVID-19 and flu.
Surveillance of Zoonotic Reservoirs
Shotgun metagenomics enables sequencing of DNA from the complete microbial community in zoonotic samples, including both novel and known species. The sequencing reads can be used to understand the emergence, evolution, and transmission of AMR gene and/or species distributions.
Healthcare-Associated Infection Surveillance
NGS-based bacterial genome sequencing paired with user-friendly bioMérieux software allows comprehensive isolate discrimination and characterization.
Frequently Purchased Together
Get help finding the right infectious disease surveillance solution for your needs.
Infectious Disease Surveillance News
Illumina and Helix Collaborate on National Surveillance Infrastructure
Read MoreLeaders in COVID Surveillance: Utah Public Health Laboratory
Read MoreExtending COVID Surveillance Capabilities Beyond Big Cities
Read MoreAustralia Tracks COVID-19 Nationwide to Understand Transmission and Spread
Read MoreHow Sequencing-Based Surveillance Helps Fight COVID-19
Read MoreFeatured Genomic Surveillance Publications
Monkeypox virus multiplexed PCR amplicon sequencing (PrimalSeq) V.2.
Phylogenomic characterization and signs of microevolution in the 2022 multi-country outbreak of monkeypox virus
Genome Sequencing of Sewage Detects Regionally Prevalent SARS-CoV-2 Variants
Genomic surveillance at scale is required to detect newly emerging strains at an early timepoint
Discovery of Bat Coronaviruses through Surveillance and Probe Capture-Based NGS
Related Solutions
Coronavirus Sequencing
NGS can characterize SARS-CoV-2 and other respiratory pathogens, identify novel coronaviruses, and more. Compare NGS methods for common coronavirus studies.
Coronavirus Software
Software tools to detect and identify viral sequences, examine COVID-19 host responses, and contribute findings to shared databases.
COVID-19 Host Genetics & Immune Response Profiling
Methods to interrogate host genetic variation and profile the molecular mechanisms that drive immune responses.
References
- Woolhouse ME, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg Infect Dis. 2005;11:1842–7. 10.3201/eid1112.050997
- Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, et al. Global trends in emerging infectious diseases. Nature. 2008;451:990–3.