
NovaSeq X Series ordering
Advanced chemistry, optics, and informatics combine to deliver exceptional sequencing speed and data quality, outstanding throughput, and scalability.
These kits provide a simple, cost-effective solution for analysis of the coding transcriptome, with minimal hands-on time.
Assay time
Hands-on time
Input quantity
Generate mRNA-focused sequencing libraries from total RNA, with enhanced multiplex capability and a simple workflow with master-mixed reagents.
Kits feature 24 unique indexes, delivering enhanced multiplex performance for processing large numbers of samples. The kits include adapters containing unique index sequences that are ligated to sample fragments at the beginning of the library construction process. This allows the samples to be pooled and then individually identified during downstream analysis.
Master-mixed reagents eliminate the majority of pipetting steps and reduce the amount of clean-up, as compared to previous methods, minimizing hands-on time. This results in economical, high-throughput RNA sequencing studies achieved with a user-friendly workflow.
Prepare libraries for Illumina sequencing in less than a day. Illumina Qualified Methods are available on a range of automation platforms through our partners.
| Assay time | ~10.5 hr |
|---|---|
| Automation capability | Liquid handling robot(s) |
| Automation details | Explore available automation methods |
| Content specifications | Captures the coding transcriptome (without strand information) |
| Description | A simple, cost-effective research solution for analysis of the coding transcriptome. |
| Hands-on time | ~4.5 hr |
| Input quantity | 0.1 - 1 ug total RNA or 10 - 400 ng previously isolated mRNA (from species with polyA tails) |
| Instruments | MiSeq System, NextSeq 550 System, NextSeq 500 System |
| Mechanism of action | Oligo-dT beads capture polyA tails |
| Method | mRNA sequencing |
| Multiplexing | Up to 24-plex per lane |
| Nucleic acid type | RNA |
| Specialized sample types | Not FFPE-compatible |
| Species category | Mammalian, Bovine, Mouse, Human, Rat |
| Species details | Works with high-quality RNA from any species with polyA tails |
| Strand specificity | Non-stranded |
| Technology | Sequencing |
| Variant class | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants |
TruSeq RNA Library Prep Kit v2
| Instrument | Recommended number of samples | Read length |
|---|---|---|
| NextSeq 550 System | RNA profiling: 13–40 samples per run (based on 10 million reads per sample) |
2 x 75 bp |
RNA-Seq uses next-generation sequencing to analyze expression across the transcriptome, enabling scientists to detect known or novel features and quantify RNA.
mRNA sequencing (mRNA-Seq) using NGS can identify both known and novel transcripts, and measure transcript abundance.
Learn how to profile gene expression changes for a deeper understanding of biology.
| TruSeq RNA Library Prep Kit v2 | Illumina Stranded mRNA Prep | Illumina RNA Prep with Enrichment | |
|---|---|---|---|
| Assay time | ~10.5 hr | 6.5 hr | < 9 hr |
| Automation capability | Liquid handling robot(s) | Liquid handling robot(s) | Liquid handling robot(s) |
| Automation details | Explore available automation methods | Explore available automation methods | |
| Content specifications | Captures the coding transcriptome (without strand information) | Captures the coding transcriptome with strand information | Captures the coding transcriptome when used with Illumina Exome Panel |
| Description | A simple, cost-effective research solution for analysis of the coding transcriptome. | A simple, cost-effective solution for analysis of the coding transcriptome with precise strand information | A reproducible, economical solution enabling targeted transcript detection and discovery from a broad range of sample types and inputs including formalin-fixed, paraffin- embedded (FFPE) tissues and other low-quality samples |
| Hands-on time | ~4.5 hr | < 3 hr | < 2 hr |
| Input quantity | 0.1 - 1 ug total RNA or 10 - 400 ng previously isolated mRNA (from species with polyA tails) | 25-1000 ng standard-quality total RNA | 10 ng RNA; 20 ng FFPE RNA |
| Instruments | MiSeq System, NextSeq 550 System, NextSeq 500 System | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System | MiSeq System, iSeq 100 System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiniSeq System, NovaSeq 6000 System, MiSeq i100 Plus System, MiSeq i100 System |
| Mechanism of action | Oligo-dT beads capture polyA tails | PolyA capture, ligation-based addition of adapters and indexes | Bead-linked transposome |
| Method | mRNA sequencing | mRNA sequencing | mRNA sequencing, Targeted RNA sequencing, Target enrichment |
| Multiplexing | Up to 24-plex per lane | Up to 384 Unique Dual Indexes (UDIs) | Up to 384 Unique Dual Indexes (UDIs) |
| Nucleic acid type | RNA | RNA | RNA |
| Specialized sample types | Not FFPE-compatible | Not FFPE-compatible, Low-input samples | Blood, Low-input samples, FFPE tissue |
| Species category | Mammalian, Bovine, Mouse, Human, Rat | Mammalian, Bovine, Mouse, Human, Rat | Human, Virus |
| Species details | Works with high-quality RNA from any species with polyA tails | Works with high-quality RNA from any species with polyA tails | |
| Strand specificity | Non-stranded | Stranded | Non-stranded |
| Technology | Sequencing | Sequencing | Sequencing |
| Variant class | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants |
Library prep and array kit selector
Find the right sequencing library preparation kit or microarray for your needs. Filter by method, species, and more. Compare, share, and order kits.
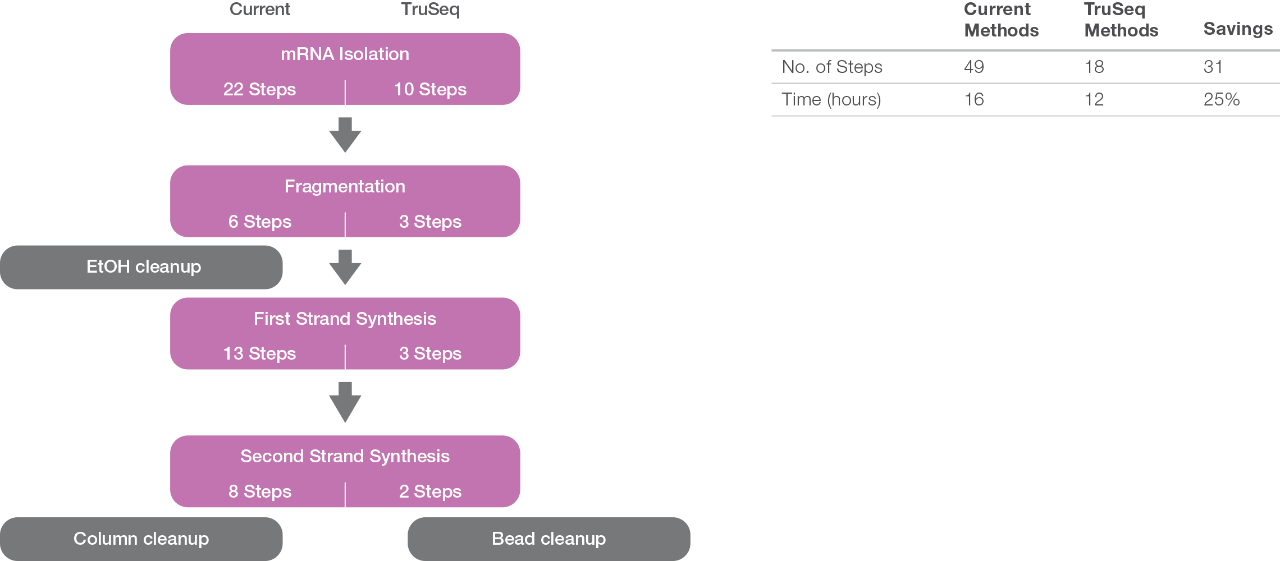

Compared to current methods for preparing mRNA samples for sequencing, use of the TruSeq reagents significantly reduces the number of steps and hands-on time.
TruSeq RNA Library Preparation Kit v2, Set A (48 samples, 12 indexes)
RS-122-2001
Includes reagents and 12 of 24 indexes (Indexes: 2, 4, 5, 6, 7, 12, 13, 14, 15, 16, 18, 19) for preparing 48 total RNA samples.
List Price:
Discounts:
TruSeq RNA Library Preparation Kit v2, Set B (48 samples, 12 indexes)
RS-122-2002
Includes reagents and 12 of 24 indexes (Indexes: 1, 3, 8, 9, 10, 11, 20, 21, 22, 23, 25, 27) for preparing 48 total RNA samples.
List Price:
Discounts:
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.