
NovaSeq X Series ordering
Advanced chemistry, optics, and informatics combine to deliver exceptional sequencing speed and data quality, outstanding throughput, and scalability.
Provides a reproducible, economical solution for sequencing RNA from FFPE tissues and other low-quality samples. Accuracy from as little as 10 ng total RNA.
Assay time
Hands-on time
Input quantity
The product previously known as the TruSeq RNA Access Library Prep Kit (Cat. No. RS-301-2001 and RS-301-2002) is now called TruSeq RNA Exome. The product configuration has changed. In the new configuration, major components such as library preparation, index adapters, enrichment reagents, and exome panels can be purchased separately. If you were previously buying Cat. No. RS-301-2001 or Cat. No. RS-301-2002, purchase the following components: TruSeq RNA Library Prep for Enrichment (Cat. No. 20020189), TruSeq RNA Enrichment (Cat. No. 20020490), Exome Panel (Cat. No. 20020183), and one item from the Index Adapters section below.
TruSeq RNA Exome, previously known as the TruSeq RNA Access Library Prep Kit, converts total RNA into template molecules of known strand origin, followed by sequence-specific capture of coding RNA. This provides a low-cost solution for analyzing human RNA isolated from formalin-fixed, paraffin-embedded (FFPE) tissues and other low-quality samples.
TruSeq RNA Exome generates RNA sequencing (RNA-Seq) libraries from degraded samples that focus on the RNA coding regions. Isolating these high-value content regions to help maximize discovery power, while requiring only a fraction of the read depth of total RNA sequencing. The results are low input requirements, high sample throughput, and cost-effective transcriptome analysis. Reagent volumes supplied are sufficient to support 1- to 4-plex enrichment reactions.
| Assay time | ~2 days |
|---|---|
| Automation capability | Liquid handling robot(s) |
| Automation details | Explore available automation methods |
| Content specifications | Captures the coding transcriptome/RNA exome |
| Description | Gives researchers a clear, comprehensive view of the coding transcriptome with precise strand information and a reduced sample input requirement. |
| Hands-on time | ~11 hr |
| Input quantity | 10 ng total RNA from fresh/frozen samples, or 20 ng total RNA from FFPE samples |
| Instruments | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System |
| Mechanism of action | Biotinylated capture probes that target coding RNA. Does not require RNA with poly-A tails. |
| Method | Exome sequencing, mRNA sequencing |
| Multiplexing | Up to 24 single, 96 combinatorial (CD) dual |
| Nucleic acid type | RNA |
| Specialized sample types | Low-input samples, FFPE tissue |
| Species category | Human |
| Strand specificity | Stranded |
| Technology | Sequencing |
| Variant class | Single nucleotide polymorphisms (SNPs), Novel transcripts, Transcript variants |
Illumina now offers modular product ordering to enable flexibility in your workflows. Please order library prep, enrichment, index adapters, and exome panel components separately.
| Instrument | Recommended number of samples | Read length |
|---|---|---|
| NextSeq 550 System | 5 to 16 samples per run (based on 25 million reads per sample) |
2 x 75 bp |
| NovaSeq 6000 System | Samples per run (dual flow cell): S1: 96, S2: 96, S4: 192 (currently limited by available index combinations) |
2 × 100 bp |
| NovaSeq 6000 System | Samples per run (dual flow cell): S1: 96, S2: 96, S4: 192 (currently limited by available index combinations) |
2 × 100 bp |
Low-Quality/FFPE RNA-Seq Solutions
Find FFPE RNA sequencing solutions that yield high-quality results from degraded FFPE tissues and other low-quality samples.
Detect cancer gene expression and transcriptome changes and identify novel cancer transcripts with RNA next-generation sequencing (RNA-Seq).
Library Prep and Array Kit Selector
Find the right sequencing library preparation kit or microarray for your needs. Filter by method, species, and more. Compare, share, and order kits.
| TruSeq RNA Exome | Illumina RNA Prep with Enrichment | Illumina Stranded mRNA Prep | |
|---|---|---|---|
| Assay time | ~2 days | < 9 hr | 6.5 hr |
| Automation capability | Liquid handling robot(s) | Liquid handling robot(s) | Liquid handling robot(s) |
| Automation details | Explore available automation methods | Explore available automation methods | |
| Content specifications | Captures the coding transcriptome/RNA exome | Captures the coding transcriptome when used with Illumina Exome Panel | Captures the coding transcriptome with strand information |
| Description | Gives researchers a clear, comprehensive view of the coding transcriptome with precise strand information and a reduced sample input requirement. | A reproducible, economical solution enabling targeted transcript detection and discovery from a broad range of sample types and inputs including formalin-fixed, paraffin- embedded (FFPE) tissues and other low-quality samples | A simple, cost-effective solution for analysis of the coding transcriptome with precise strand information |
| Hands-on time | ~11 hr | < 2 hr | < 3 hr |
| Input quantity | 10 ng total RNA from fresh/frozen samples, or 20 ng total RNA from FFPE samples | 10 ng RNA; 20 ng FFPE RNA | 25-1000 ng standard-quality total RNA |
| Instruments | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System | MiSeq System, iSeq 100 System, NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, MiniSeq System, NovaSeq 6000 System, MiSeq i100 Plus System, MiSeq i100 System | NextSeq 550 System, NextSeq 2000 System, NextSeq 1000 System, NovaSeq X System, NextSeq 500 System, NovaSeq 6000 System, NovaSeq X Plus System |
| Mechanism of action | Biotinylated capture probes that target coding RNA. Does not require RNA with poly-A tails. | Bead-linked transposome | PolyA capture, ligation-based addition of adapters and indexes |
| Method | Exome sequencing, mRNA sequencing | mRNA sequencing, Targeted RNA sequencing, Target enrichment | mRNA sequencing |
| Multiplexing | Up to 24 single, 96 combinatorial (CD) dual | Up to 384 Unique Dual Indexes (UDIs) | Up to 384 Unique Dual Indexes (UDIs) |
| Nucleic acid type | RNA | RNA | RNA |
| Specialized sample types | Low-input samples, FFPE tissue | Blood, Low-input samples, FFPE tissue | Not FFPE-compatible, Low-input samples |
| Species category | Human | Human, Virus | Mammalian, Bovine, Mouse, Human, Rat |
| Strand specificity | Stranded | Non-stranded | Stranded |
| Technology | Sequencing | Sequencing | Sequencing |
| Variant class | Single nucleotide polymorphisms (SNPs), Novel transcripts, Transcript variants | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants | Single nucleotide polymorphisms (SNPs), Gene fusions, Novel transcripts, Transcript variants |
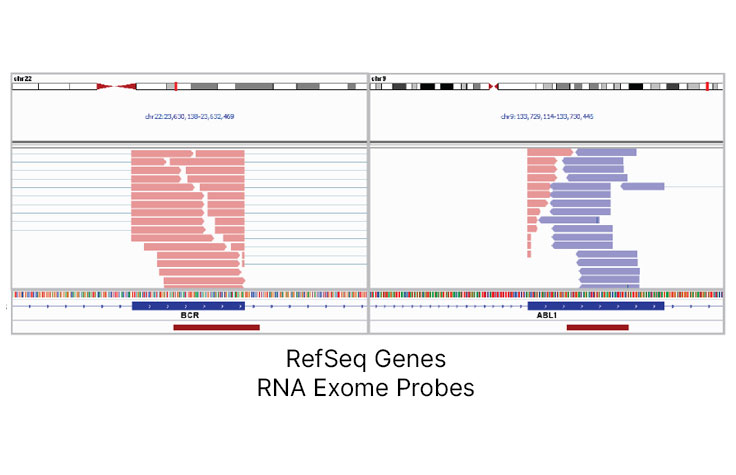

Efficient Gene Fusion Discovery
TruSeq RNA Exome enables detection of expressed fusion transcripts without the need to design probes specific for the fusion junction. The well-characterized BCR-ABL fusion is detected efficiently in the Universal Human Reference RNA (UHRR) sample at 25 M reads.
Ebola and sexual transmission
Six months after being discharged from the Ebola treatment, a male survivor of Ebola Virus Disease (EVD) transmitted the virus to his female partner, who died of the disease. The survivor’s semen had tested negative by conventional means, but the authors were able to enrich the semen sample for EBOV RNA with the Illumina TruSeq RNA Access Kit and custom probes.
TruSeq® RNA Library Prep for Enrichment (48 Samples)
20020189
Includes reagents for preparing 48 libraries. Purchase index adapters, enrichment panel, and enrichment reagents separately.
List Price:
Discounts:
TruSeq® RNA Enrichment (12 enrichments)
20020490
Sufficient for 12 enrichment reactions when paired with TruSeq RNA Library Prep for Enrichment. Purchase library prep, index adapters, and probe panels separately.
List Price:
Discounts:
TruSeq RNA Single Indexes Set A (12 Indexes, 48 Samples)
20020492
Includes 12 of 24 indexes (Indexes: 2, 4, 5, 6, 7, 12, 13, 14, 15, 16, 18, 19) for preparing 48 individual RNA samples.
List Price:
Discounts:
TruSeq RNA Single Indexes Set B (12 Indexes, 48 Samples)
20020493
Includes 12 of 24 indexes (Indexes: 1, 3, 8, 9, 10, 11, 20, 21, 22, 23, 25, 27) for preparing 48 individual RNA samples.
List Price:
Discounts:
Illumina Exome Panel – Enrichment Oligos Only
20020183
Sufficient for 8 enrichment reactions when paired with Illumina DNA Prep with Enrichment, 12 reactions when paired with TruSeq RNA Exome, and 32 reactions when paired with Illumina RNA prep with enrichment. Purchase library prep and enrichment reagents and index adapters separately.
List Price:
Discounts:
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.