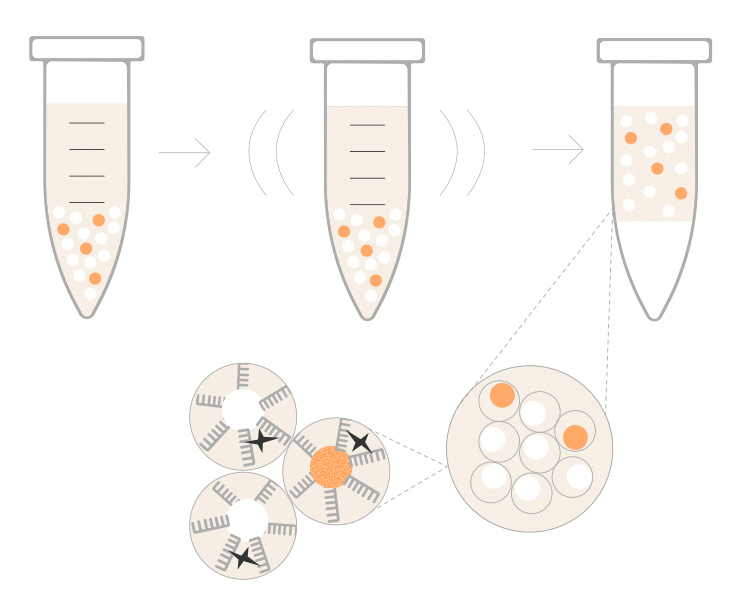
Revolutionizing single-cell sequencing
Download the eBook to explore Illumina Single Cell 3′ RNA Prep chemistry. Learn how it reduces costs, simplifies workflows, and delivers high-quality libraries for a more complete view of your tissue.
Revolutionizing single-cell sequencing
Your email address is never shared with third parties.