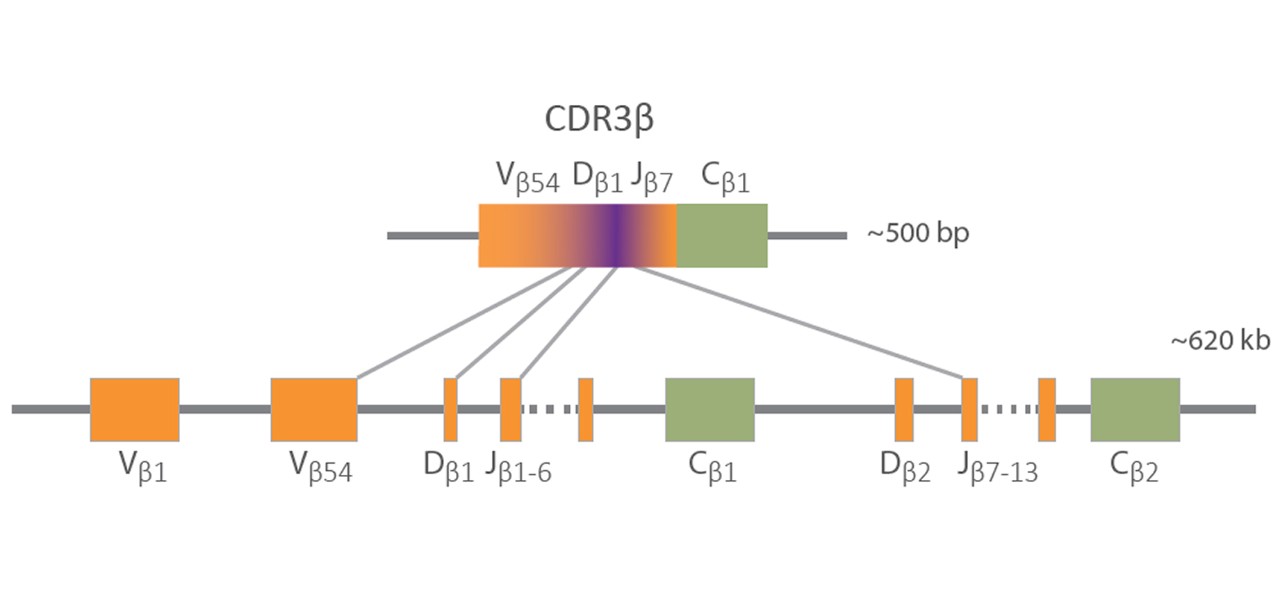
Multiomics is an integrated approach that provides powerful biological insights by combining data sets from multiple molecular levels, including genomics, transcriptomics, epigenetics, and proteomics. Multiomics enables a more holistic and integrated understanding of how different biological levels interact. Furthermore, the combination of different NGS techniques can provide multiomic insights that expand our understanding of the immune repertoire, drug resistance mechanisms, and possible future disease treatment strategies.6
Multiomics data sets are inherently complex, making analysis a significant challenge. Illumina Connected Multiomics software empowers researchers with intuitive, scalable analysis, for streamlined sample-to-insights workflows. With these built-in capabilities, biologists can confidently generate powerful statistics and interactive visualizations without a bioinformatics background. Explore our multiomics resource page to expand your research across multiple omes and learn about Illumina Connected Multiomics software for multiomics data analysis and visualization needs.