
NovaSeq X Series ordering
Advanced chemistry, optics, and informatics combine to deliver exceptional sequencing speed and data quality, outstanding throughput, and scalability.
Error correction with unique molecular identifiers (UMIs) for next-generation sequencing libraries.
Assay time
Hands-on time
Input quantity
These reagents reduce background noise in sequencing data, enabling detection of low-frequency variants, such as those found in cell-free DNA (cfDNA).
Circulating tumor DNA (ctDNA) may represent a very small fraction of cfDNA, near the limit of detection for next-generation sequencing (NGS). The TruSight Oncology UMI Reagents address this challenge with UMIs and error correction software, reducing error rates to < 0.007% and enabling detection of low-frequency variants. Lower error rates increase analytical specificity, resulting in higher confidence in NGS data.
The TruSight Oncology UMI Reagents include UMI adapters and indexes, plus DNA library prep and enrichment reagents. These reagents can also be paired with the TruSight Tumor 170 DNA oligos. Integration of UMIs does not create any extra steps in the library prep workflow.
The UMI Error Correction App aligns reads, then uses UMIs to exclude false positives, reducing variant calling errors. The UMI Error Correction App is available in the cloud-based BaseSpace Sequence Hub or for local installation.
| Assay time | 1.5 days |
|---|---|
| Cancer type | Pan-cancer |
| Hands-on time | 5 hr |
| Input quantity | 30 ng cell-free DNA |
| Instruments | NextSeq 550 System, NextSeq 2000 System, NovaSeq 6000 System |
| Method | Targeted DNA sequencing |
| Nucleic acid type | DNA |
| Specialized sample types | Not FFPE-compatible, Cell-free DNA |
| Species category | Human |
| Technology | Sequencing |
TruSight Oncology UMI Reagents do not contain any enrichment probes. If you wish to use the TruSight Tumor 170 DNA probe set, purchase the TruSight Tumor 170 Content Set separately.
For convenience, the TruSight cfDNA instrument bundles contain all reagents necessary to generate libraries and sequence 48 samples at the recommended sequencing depth for cell-free DNA (cfDNA) libraries on the selected instrument.
TruSight Oncology UMI Reagents
Local UMI Error Correction App
Contact your local Illumina Support team to access the Local UMI Error Correction App.
* Performance on the NovaSeq 6000 System was demonstrated but not extensively tested.
Pathology and clinical cancer research
Our clinical cancer research solutions deliver accurate genomic information, and enable labs to analyze multiple genes in a single test.
With targeted sequencing or resequencing, a subset of genes or a genomic region of interest is isolated and sequenced, which can conserve lab resources.
NGS-based cancer sequencing methods provide more information in less time compared to single-gene and array-based approaches.
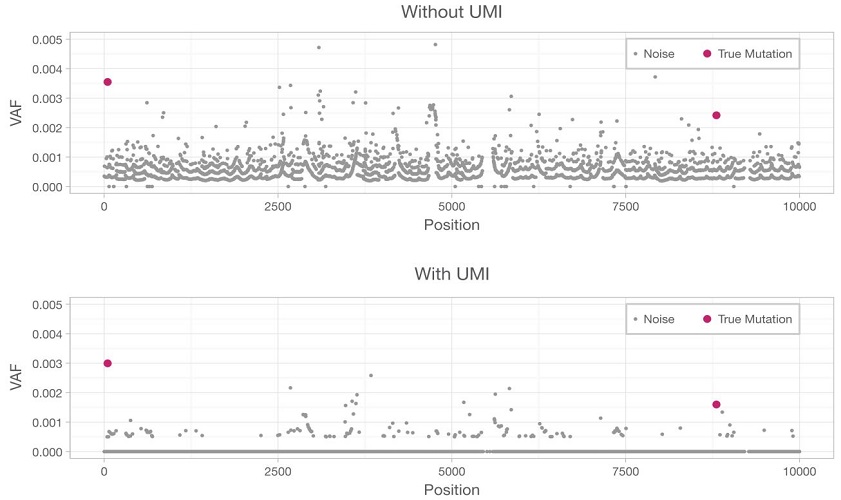

To enable accurate detection of rare variants, UMIs are integrated with error correction software, enabling true mutations to be distinguished from background noise. The bottom panel illustrates how, by removing inherent errors that result in false positives, the reduced error rate allows true mutations (red dots) to be better distinguished from background noise (grey dots).
Library preparation was performed using the TruSight Oncology UMI Reagents paired with DNA content from the TruSight Tumor 170 DNA assay. 31 samples were distributed among four independent sequencing runs on the HiSeq 4000 System. Mean error rates are shown with and without collapsed reads using the UMI Error Correction App.
| Sequencing Run | Mean Error Rate (Uncollapsed Reads) | Mean Error Rate (Collapsed Reads) |
|---|---|---|
| 1 | 0.038% | 0.0023% |
| 2 | 0.043% | 0.0024% |
| 3 | 0.035% | 0.0024% |
| 4 | 0.084% | 0.0019% |
TruSight Oncology UMI Reagents (16 indexes, 48 samples)
20024586
Contains the TruSight Oncology DNA library prep and enrichment reagents plus the TruSight UMI Toolkit. Provides sufficient reagents for the generation of 48 UMI-containing libraries with 16 unique index pairs. Utilized to generate libraries with reduced error rates, enabling the detection of low frequency variants. Contains no enrichment probes - TST170 content may be purchased separately.
List Price:
Discounts:
TruSight® Tumor 170 Content Set
20010188
DNA and RNA probes for 170 genes in TST170 kit.
List Price:
Discounts:
Showing of
Product
Qty
Unit price
Product
Catalog ID
Quantity
Unit price
Reach out for information about our products and services, or get answers to questions about our technology.
Your email address is never shared with third parties.